For the common identification method of a single protein, we must immediately think of Western Blot. Western Blot is to identify the target protein by the specific binding of the antibody and the target molecule, but this method is suitable for the identification of known proteins and must rely on antibodies, so it is not suitable for unknown or no corresponding antibody. analysis. So the little friends may ask, what if the protein you want to study is unknown or there is currently no available antibody? In this case, the protein can be identified by means of mass spectrometry identification technology. In this issue, the editor has sorted out the relevant knowledge of protein identification for everyone, hoping to help students who have been confused about protein identification.
Single protein isolation and purification
Referring to a single protein sample, as the name implies, this type of protein sample should have undergone certain protein purification steps to obtain a relatively single protein. Usually, the proteins synthesized by cell lysis or in vitro contain other miscellaneous proteins, so it is necessary to distinguish the target protein from the miscellaneous proteins. The commonly used protein separation and purification method is electrophoresis separation. One-dimensional electrophoresis separation utilizes the characteristics of different protein sizes, which can well distinguish proteins of different molecular weights. In addition, on the basis of two-dimensional electrophoresis, protein molecules with different isoelectric points can be further distinguished.
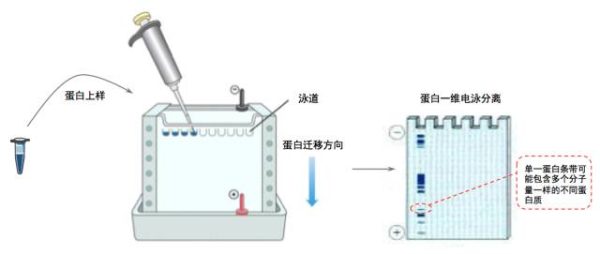
one-dimensional electrophoresis
The one-dimensional electrophoresis commonly used at present is vertical plate polyvinylamide gel electrophoresis (SDS-PAGE gel). The SDS surfactant neutralizes the charge on the surface of the protein. When the denatured protein mixed sample passes through the gel gap, the proteins of different molecular weights migrate at different speeds in the gel and are thus separated. One-dimensional electrophoresis enables the separation of most proteins and protein complexes for initial separation.
two-dimensional electrophoresis
Two-dimensional electrophoresis, also known as two-dimensional electrophoresis, is based on one-dimensional electrophoresis with the addition of lateral isoelectric focusing separation, and its vertical separation is exactly the same as that of one-dimensional SDS separation. Two-dimensional electrophoresis first uses the different characteristics of protein isoelectric points to perform preliminary separation of proteins, and then further conducts two-dimensional separation of proteins with the same or similar isoelectric points according to different protein molecular weights to further improve the protein separation efficiency. Compared with one-dimensional electrophoresis, the protein resolution of two-dimensional electrophoresis has been greatly improved. After two-dimensional electrophoresis separation, most proteins can be separated into single protein gel spots.
single protein identification
The current methods for the identification of higher purity or single proteins include protein fingerprinting (PMF) and MS/MS tandem mass spectrometry. In both methods, the protein enzyme is first cut into peptide fragments, and then the peptide fragments are analyzed by mass spectrometer to achieve the purpose of protein identification. The difference between the two methods is that the PMF method only uses primary mass spectrometry to detect protein peptides, while MS/MS not only undergoes primary mass spectrometry detection, but also selects specific peptides for peptide fragmentation on this basis. The sequence of the peptide was determined. Therefore, MS/MS has higher identification accuracy than PMF and higher reliability compared with the database, but the analysis is also more complicated.
Protein Fingerprinting (PMF)
Due to the difference in protein sequence, the size and quality of the polypeptide fragments produced by the digestion of different proteins under the action of the same digestive enzyme are specific. The protein fingerprint identification method takes advantage of the different characteristics of the fragments produced by each protease cleavage, digests the purified protein, and then analyzes the quality of the peptide fragments produced by the cleavage to obtain the peptide quality of the protein. Spectrum. Then, the peptide map is compared with the theoretical peptide map of the protein in the database, and then the identity information of the protein can be obtained.
MS/MS tandem mass spectrometry
The principle of tandem mass spectrometry (MS/MS) for protein detection is to first use primary mass spectrometry to detect the mass information of peptides, and then select peptides with high peaks for fragmentation, and analyze to obtain secondary mass spectrometry. Use the search software to select the corresponding database to analyze the mass spectrometry data, and at the same time judge the identification results in the form of scoring.
Protein Mixture Identification
Although there are many methods for protein separation and purification, the separation efficiency is not high enough in many cases, resulting in the separated protein may still contain multiple proteins. In addition, proteins with lower abundance may cause further losses during the separation process and cannot be identified, so it is necessary to directly analyze this type of proteins without separation. At present, the method suitable for the analysis of mixed proteins is LC-MS/MS chromatography-mass spectrometry tandem analysis. Compared with PMF method and MS/MS method, LC-MS/MS adds liquid chromatography separation before mass spectrometry analysis, which makes the protein analysis more precise, and the peptide sequence determination can reach almost 100% accuracy. Therefore, the number of proteins that can be identified by this service is also higher than that of individual protein tapes. LC-MS/MS can be used to identify protein mixture samples with about 200 proteins, including immunoprecipitation samples, Pull-down samples, etc.
